Assessment of Binding Affinity via Alchemical Free-Energy Calculations | Journal of Chemical Information and Modeling
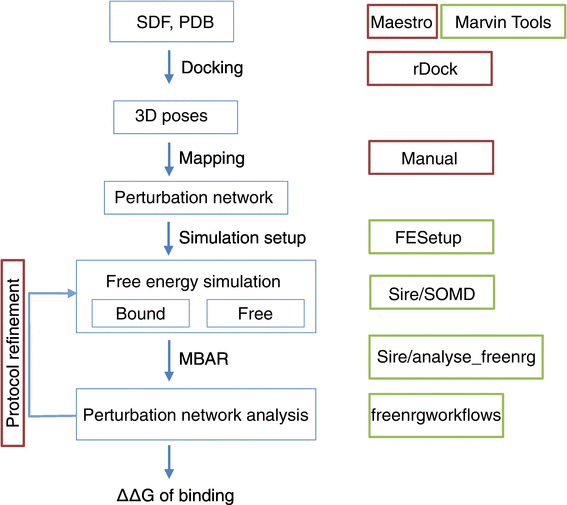
Impact of domain knowledge on blinded predictions of binding energies by alchemical free energy calculations | SpringerLink
CHEM5302 Fall 2017: Docking and BEDAM Free Energy Calculations for LEDGF Inhibitors of HIV Integrase

REST-MD visualizations, implemented in Maestro (Schrödinger, 2020b)... | Download Scientific Diagram
a) Maestro panels illustrating the receptor grid generation procedure.... | Download Scientific Diagram
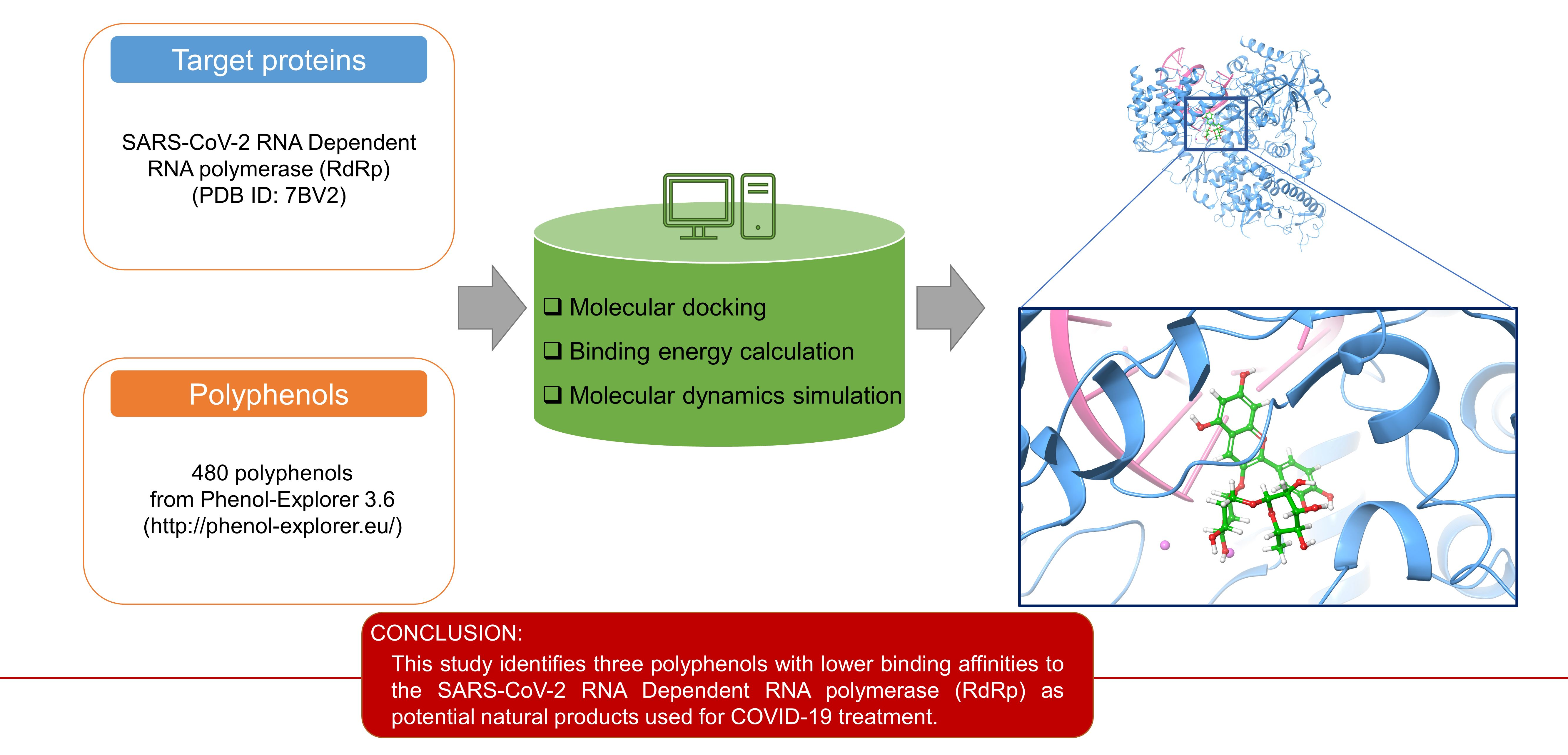
Molecules | Free Full-Text | Polyphenols as Potential Inhibitors of SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp)
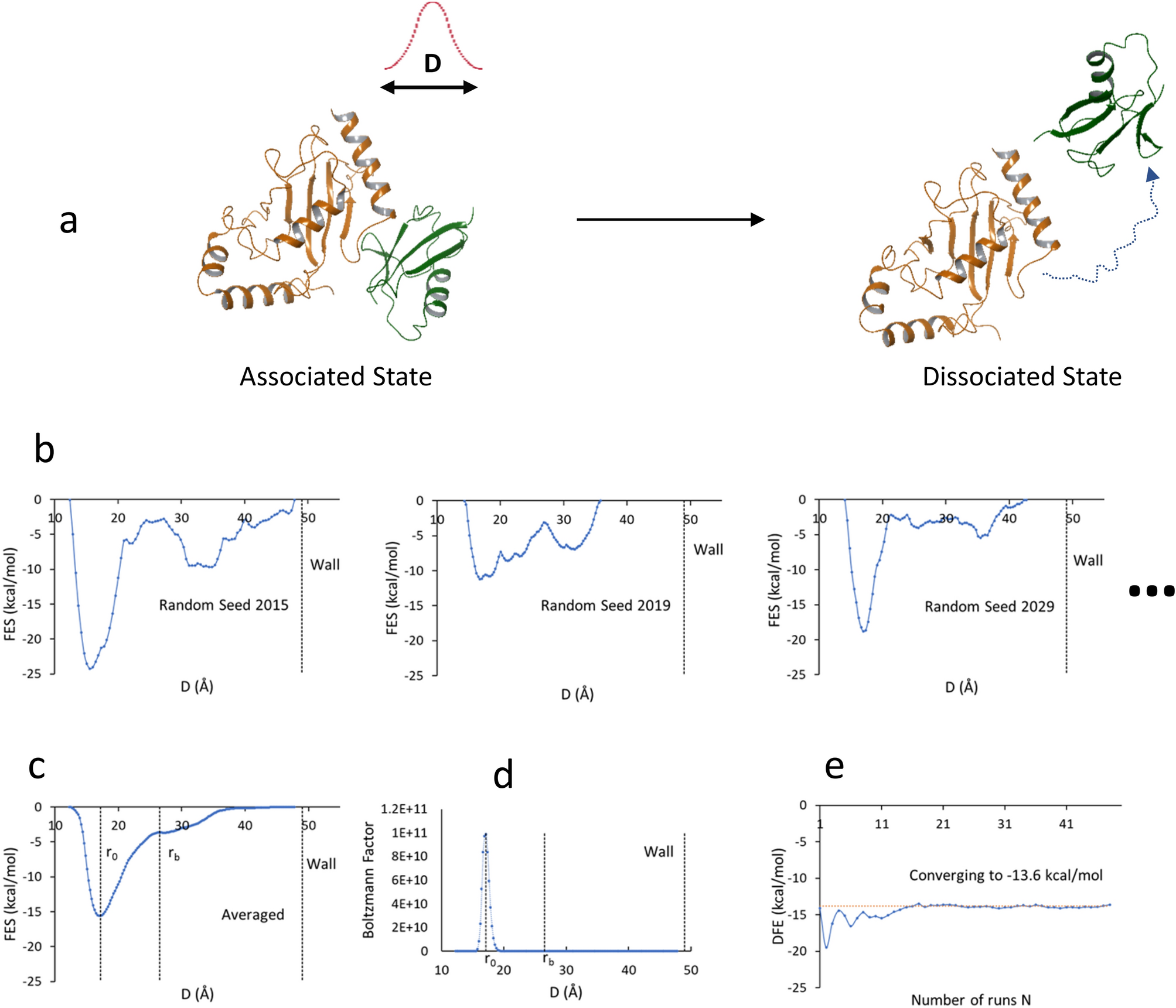
A highly accurate metadynamics-based Dissociation Free Energy method to calculate protein–protein and protein–ligand binding potencies | Scientific Reports

Polymers | Free Full-Text | The Anti-Inflammatory Effect of Lactose-Modified Hyaluronic Acid Molecules on Primary Bronchial Fibroblasts of Smokers

Frontiers | Relative binding free energy calculations with transformato: A molecular dynamics engine-independent tool